ATP5A qPCR Primer Design Guide for Accurate Gene Expression Analysis

Accurate gene expression analysis is crucial for understanding biological processes, disease mechanisms, and therapeutic responses. One key gene often studied is ATP5A, which encodes a subunit of the ATP synthase complex involved in mitochondrial energy production. Designing effective qPCR primers for ATP5A ensures reliable and reproducible results. This guide provides step-by-step instructions for ATP5A qPCR primer design, focusing on best practices to optimize accuracy and efficiency in your experiments, (qPCR primer design, gene expression analysis, ATP5A gene, mitochondrial function).
Understanding ATP5A and Its Role in Gene Expression Analysis
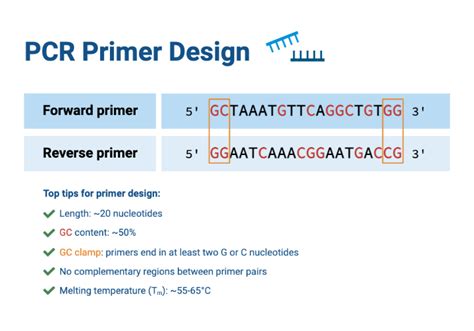
The ATP5A gene is essential for oxidative phosphorylation, making it a critical target in studies related to metabolism, cancer, and neurodegenerative diseases. Quantitative PCR (qPCR) is the gold standard for measuring gene expression due to its sensitivity and specificity. However, the success of qPCR relies heavily on well-designed primers. Poor primer design can lead to non-specific amplification, reduced efficiency, or inaccurate results, (ATP synthase, oxidative phosphorylation, qPCR efficiency).
Key Considerations for ATP5A qPCR Primer Design
- Specificity: Ensure primers bind only to the ATP5A transcript to avoid cross-reactivity with other genes or isoforms.
- Amplicon Length: Aim for 100–200 bp to optimize amplification efficiency and compatibility with qPCR platforms.
- GC Content: Maintain 40–60% GC content to ensure stable primer binding and avoid secondary structures.
- Tm Matching: Design primers with similar melting temperatures ™ to ensure uniform annealing during PCR cycles.
Step-by-Step Guide to ATP5A qPCR Primer Design
Follow these steps to design high-quality ATP5A qPCR primers:
Step 1: Retrieve ATP5A Sequence Information
Obtain the ATP5A nucleotide sequence from reliable databases such as NCBI GenBank or Ensemble. Verify the sequence to ensure it corresponds to the specific isoform or variant of interest, (NCBI GenBank, Ensemble database, nucleotide sequence).
Step 2: Select Primer Design Tools
Use specialized software like Primer3, Primer-BLAST, or Thermo Fisher’s OligoPerfect for primer design. These tools consider factors like Tm, GC content, and secondary structure prediction, (Primer3, Primer-BLAST, OligoPerfect).
Step 3: Define Primer Parameters
Set the following parameters for optimal primer design:
| Parameter | Recommended Range |
|---|---|
| Primer Length | 18–25 bp |
| Tm | 58–62°C |
| GC Content | 40–60% |
| Amplicon Length | 100–200 bp |

💡 Note: Avoid designing primers across exon-exon junctions unless splicing variants are being studied.
Step 4: Validate Primer Specificity
Use BLAST to confirm primer specificity and avoid off-target binding. Ensure the primers align only to the ATP5A sequence and not to other genes or pseudogenes, (BLAST search, primer specificity, off-target binding).
Checklist for ATP5A qPCR Primer Design

- Retrieve the correct ATP5A sequence from a reliable database.
- Use primer design tools with appropriate parameters.
- Validate primer specificity using BLAST.
- Order primers with purification options (e.g., HPLC or cartridge) for better quality.
Designing effective ATP5A qPCR primers is essential for accurate gene expression analysis. By following this guide, you can ensure specificity, efficiency, and reproducibility in your qPCR experiments. Proper primer design not only saves time and resources but also enhances the reliability of your research findings, (qPCR primer design, gene expression analysis, ATP5A gene, mitochondrial function).
Why is primer specificity important for ATP5A qPCR?
+
Primer specificity ensures that the primers bind only to the ATP5A transcript, avoiding amplification of non-target sequences, which can lead to inaccurate results.
What is the ideal amplicon length for ATP5A qPCR primers?
+
The ideal amplicon length is between 100–200 bp, as it ensures efficient amplification and compatibility with most qPCR platforms.
How can I validate ATP5A primer specificity?
+
Use BLAST to check if the primers align only to the ATP5A sequence and not to other genes or pseudogenes.



